
Orchestrating Self-Replication in Artificial Cells through Digital Microfluidics
10.1101/2025.05.23.655734
DMF Replicator
A defining feature of living cells is their ability to self-replicate; but creating artificial cells with this capability remains challenging, due to the complexity of biological division machinery. Rather than seeking to reconstitute this machinery, here we take direct control of DNA replication and compartment division using digital microfluidics. This approach allows us to precisely orchestrate these two fundamental processes, providing insight into how they must be coupled for successful self-replication.
Group Scientific Away Day in Brighton
May, 2025

Triply Responsive Control of Anion/Cation Transport with an Artificial Ion Channel Responsive to Multiple Stimuli
10.26434/chemrxiv-2025-sbp1x
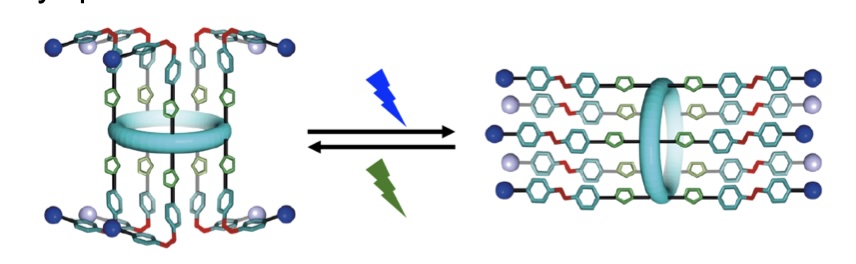
azo-p5 ion channels.
We present a synthetic ion channel whose activity can be controlled by three orthogonal stimuli (light, pH, guest/ligand). The channel is formed from a pillar[5]arene functionalized with photoswitchable tetrafluoroazobenzene moieties. We demonstrate excellent control over E:Z switching across ten incorporated photoswitches (E to Z 87%, Z to E quant.). We show that the most active isomer – the Z-isomer – forms dimeric ion channels in membranes, with selectivity for M+/Cl− symport. Single molecule planar bilayer conductance studies show distinct high and low conductance states dependent on irradiation wavelength. Finally, we demonstrate that this activity can be modulated over 170-fold by controlling pH, irradiation, and guest addition, creating a powerful addition to the canon of synthetic ion channels.
Rational Design Principles for De Novo α-Helical Peptide Barrels with Dynamic Conductive Channels
10.1021/jacs.4c13933
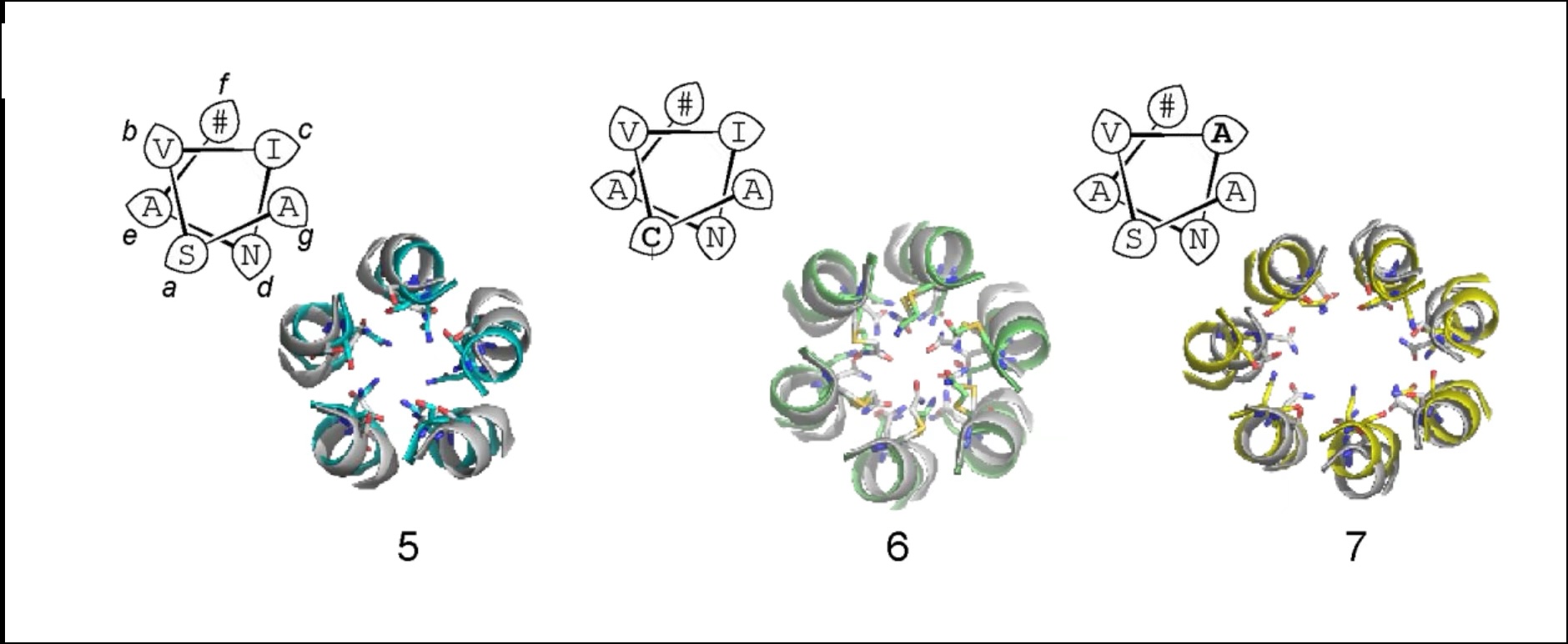
α-helical peptide channels.
Despite advances in peptide and protein design, the rational design of membrane-spanning peptides that form conducting channels remains challenging due to our imperfect understanding of the sequence-to-structure relationships that drive membrane insertion, assembly, and conductance. Here, we describe the design and computational and experimental characterization of a series of coiled coil-based peptides that form transmembrane α-helical barrels with conductive channels.
Real-time label-free imaging of living crystallization-driven self-assembly
10.1038/s41467-025-57776-9
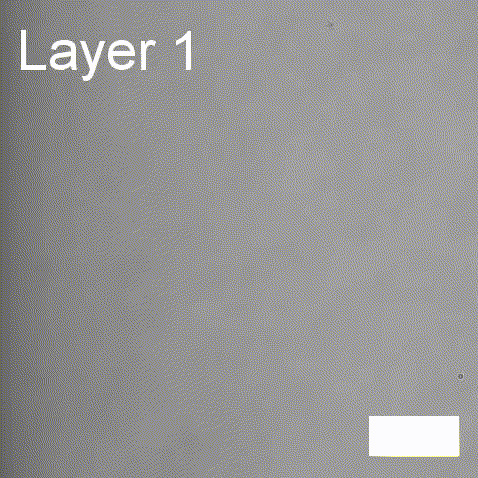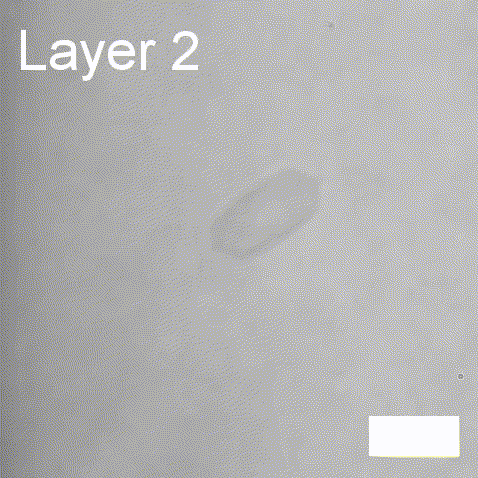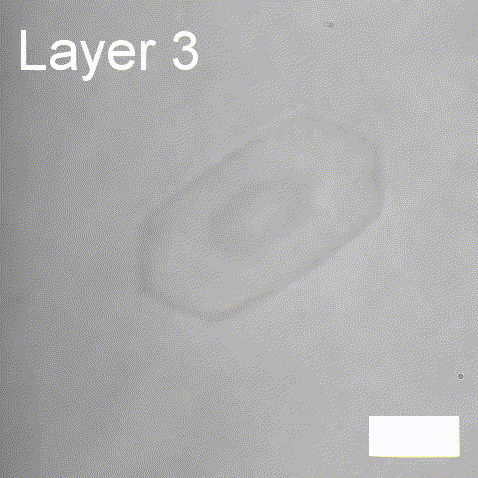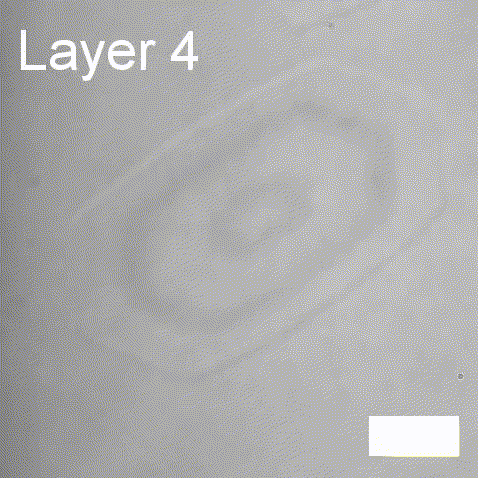
Real-time label-free imaging of living crystallization-driven self-assembly.
Our latest work now published in Nature Communications. This is the last of our pandemic papers, so no membrane biophysics! Applying label-free microscopy to watch polymer self-assembly in real-time. This lets us map directly how reaction parameters affect the composition and growth of individual 2D polymer microplatelets. A fantastic collaboration with Rachel O’Reilly at the University of Birmingham. Congratulations go to Yujie Guo for another fantastic bit of science.
Yimeng Zhu
February, 2025

Yimeng Zhu
Yimeng joined the group as a BiPAS PhD student after completing her BSc in Neuropharmacology at King’s College London and her MSc in Neuroimaging at UCL. Her project will focus on the de novo design of protein channels and their application in creating artificial synapses between synthetic and biological cells.
Sharavanakkumar Sundararajan Kasiviswanathan
February, 2025

Sharavanakkumar Sundararajan Kasiviswanathan
Sharavana completed his MSc in Biotechnology at the University of Mysore in 2022. Before beginning his PhD in February 2025, he worked on outer membrane porins from Gram-negative bacteria, studying antibiotic translocation using single-channel electrophysiology in his previous role in India. His PhD research focuses on the rational design and construction of de novo protein nanopores for single-molecule sensing applications.
Anna Worsley
February, 2025

Anna Worsley
Anna joins the group as a PhD student following completion of her Chemistry MSci at the University of Bristol. Her master’s project introduced her to the world of ultrafast spectroscopy and sparked a strong interest in the field of single-molecule imaging. In collaboration with Oxford Nanopore Technologies, she is now applying a combination of optical and electrical techniques to gain high-resolution insights into translocation events in nanopores.
Ion Transport and Membrane Channel Formation Using a Peptidomimetic in Droplet Interface Bilayers
10.1039/D4CC05926C
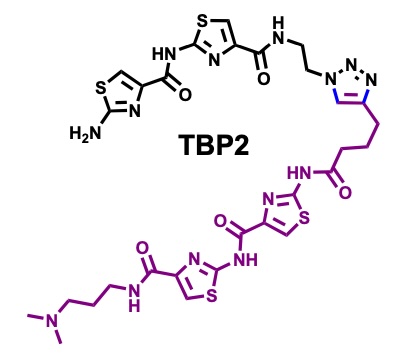
Peptidomimetic channels.
Herein, we present a synthetic peptidomimetic, TBP2, which forms artificial membrane channels within Droplet Interface Bilayers (DIBs). Through real-time electrophysiology, TIRF microscopy, and single-channel recordings, we demonstrate the ability of TBP2 to mediate high-conductance transport of Na⁺ and K⁺ across the DIBs, highlighting its potential for
nanobiotechnology applications in cell-free systems.