Dissecting the Self-Assembly Kinetics of Multimeric Pore-forming Toxins.
10.1098/rsif.2015.0762
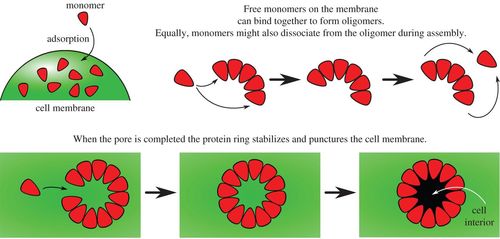
Schematic diagrams illustrating monomer addition as a proposed pathway to pore formation.
Pore-forming toxins are ubiquitous cytotoxins that are exploited by both bacteria and the immune response of eukaryotes. These toxins kill cells by assembling large multimeric pores on the cell membrane. However, a quantitative understanding of the mechanism and kinetics of this self-assembly process is lacking. We propose an analytically solvable kinetic model for stepwise, reversible oligomerization. In biologically relevant limits, we obtain simple algebraic expressions for the rate of pore formation, as well as for the concentration of pores as a function of time. Quantitative agreement is obtained between our model and time-resolved kinetic experiments of Bacillus thuringiensis Cry1Ac (tetrameric pore), aerolysin, Staphylococcus aureus α-haemolysin (heptameric pores) and Escherichia coli cytolysin A (dodecameric pore). Furthermore, our model explains how rapid self-assembly can take place with low concentrations of oligomeric intermediates, as observed in recent single-molecule fluorescence experiments of α-haemolysin self-assembly. We propose that suppressing the concentration of oligomeric intermediates may be the key to reliable, error-free, self-assembly of pores.